HPC Scripts for Global Sensitivity and Uncertainty Analysis
Download scripts, sample inputs and documentation:
- HPC scripts for UF HiperGator (SLURM HPC linux cluster) ( [1.2MB]
- HPC scripts for UCLouvain CECI cluster (SLURM HPC linux cluster) ( [4.8MB]
- HPC scripts for UPNA cluster (SGE linux cluster) ( [2.8MB]
Description
Scripts to create and manage multiple jobs needed to run a typical throughput High Performance Computing (HPC) simulation problem associated with global sensitivity and uncertainty analysis (GSUA) (see here for eSU GSA screening tool). The scripts are designed to take a sample matrix, matrix.txt (columns are input factors and rows individual samples, where the total number of rows is the total number of simulations needed for GSUA). The scripts are designed to run in an HPC linux cluster with SLURM job manager (see example for UF Hipergator HPC or UCLouvain CECI), or SGE scheduler (see example scripts above for UPNA).
Brief description of the scripts and use are presented below for the UF HiperGator with SLURM. For other SLURM or SGE implementations the instructions will be somewhat different. Please refer to the documentation of those systems for details. For additional information 4 instructional videos with step-by-step information for a typical application case are provided at the bottom of the page.
Program Usage & Output
I. Preliminaries
1. Login to you HPC account.
2. Download the zip file from the link above and transfer to your HPC home directory. Uncompress the class files (.zip) into your HPC home directory to create the class_GSA directory and its contents (notice that the first '>' in the instructions following indicates at the HPC unix command line),
> unzip -X /ufrc/abe6933/share/class_GSA.zip
3. Request an HPC development node for 2 hrs to play around without bothering others. Notice that we need to load access to the UF Research Computing tools like srundev first.
> module load ufrc > srundev -t 120
4. After the development node returns to the command line (it might take a few seconds), load the module (HPC extensions and libraries) needed to process our class problem.
> module load class/abe6933
5. If you have not done this so, your PATH (directories where programs are looked for at the command line by default, regardless on what directory you are on at that time) must include the current directory you are on ".". If you have not modified your /home/YOURGATORLINK/.bash_profile you must do so to add "." to the PATH line. This line should contain:
PATH=$PATH:.:~/bin
Notice the ":.:" in the line above. If it is not there add it to the .bash_profile with nano, vi or another unix editor of your choice and save it. When you log out and in again you will have the capability of running programs from the directory you are on by default. This is important as many unix scripts rely on finding programs where you are. If you do not want to log out but continue in the current session, you can type,
> source .bash_profile
II. Explanation of directories under class_GSA/
On the root directory class_GSA (here on called 'class directory') are the files to automate the creation and submission of HPC jobs (make_jobs.sh, class_GSA.job.top class_GSA.job.bottom, matrix.txt ). Please see subsection IV below for a brief explanation on these files. The rest of the files are organized in the foillowing subdirectories:
A) bin/ - contains unix utilities abut and linex used in sens_class unix script described below. From the command line in the class directory copy the bin/ directory to your home directory (notice that this home bin directory is always accessible based on the PATH assigned above).
> cp -Rf bin ~
B) src_java/ - source code of the java class program (not needed for the HPC simulations). This is a classical prey/predator ("Gluttons & Muttons") dynamic simulation game (Sim2D, Alberts, J., 2017). It is an object-oriented code written in java. For this example, to achieve stability of the results, the code was modified so that the population of prey (Gluttons) is set to constant (equal to the initial value) during the simulation. The program was packaged (jar) and added an option to run on the HPC linux command line without GUI (-gsua).
C) base/ - baseline execution directory for the class code. It contains:
a) PredPrey1.jar, the model executable (jar) for the HPC GSUA "Gluttons & Muttons" (see Example below).
b) README_PredPrey1.txt, description of the model inputs and outputs for the example
c) input.dat.ptr, pattern input file
d) par.txt, file with labels of input factors to change during the simulations
e) sens_class, unix script to substitute values command line vlues in model input files, run simulation and extract output of interest into matrix file outputs.all. See next section for brief explanation..
f) a_subs, unix AWK script to substitute values in input ptr files (labels must be as in par.txt)
g) test.sh, to test the substitution of inputs with your base run
h) output/: directory where results of simulations are written after jobs are done.
D) Outputs/ - this is the directory where HPC output files will be written one the jobs are completed.
III. Substitution and run script, sens_class
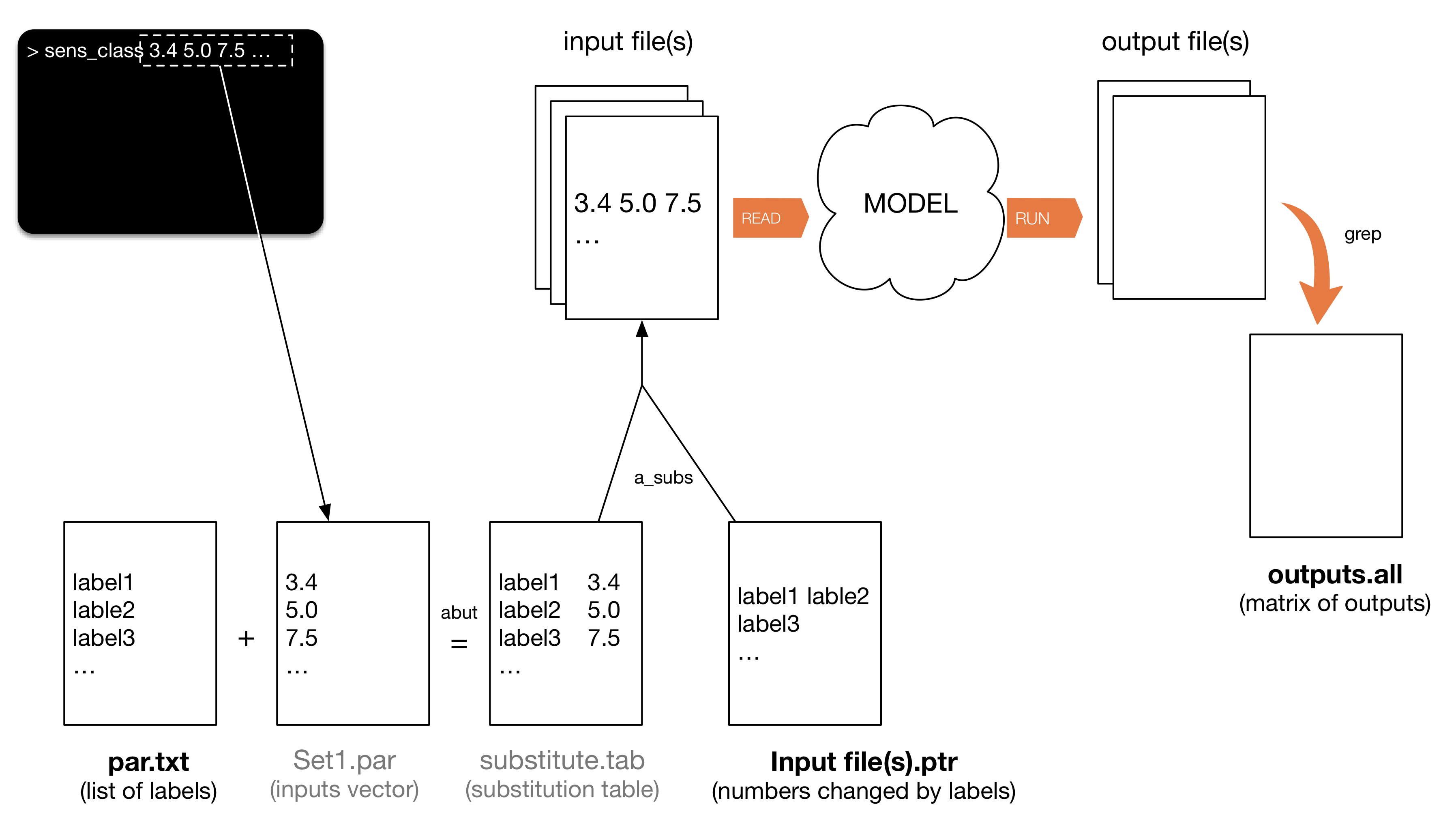
The unix script sens_class is provided to run GSUA throughput simulations, where each simulation is performed with a different input sample. The script sens_class is model independent and generic, created to substitute inputs from ASCII text input files of any model, and run the simulations. A flowchart of its behaviour and auxiliary files needed is depicted in the Figure below. Notice that file names in grey in the Figure below are temporary (deleted after each sample file is run).
IV. Automatic job creation and run script, make_jobs
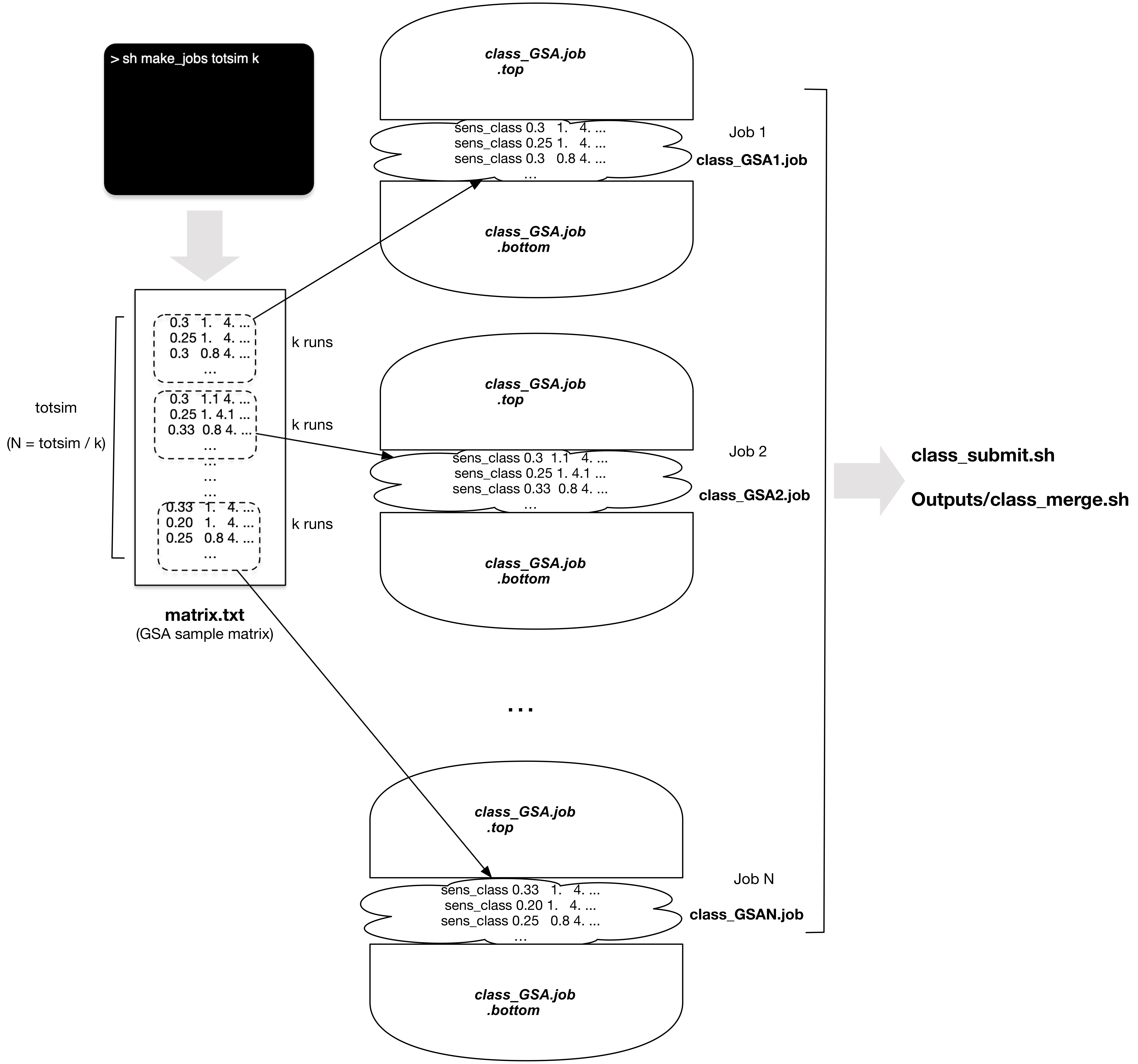
The unix script make_jobs.sh is provided to create a number of jobs from the initial GSUA sampling matrix, matrix.txt. The matrix consists of columns (input factors) and rows (sample sets). The script divides the total sumber of simulations (totsim) into blocks of k simulations each and builds an HPC SLURM job file for each block using common header and footer lines (auxiliary files class_GSA.job.top and class_GSA.job.bottom>). It then creates a script class_submit.sh to submit all jobs to the SLURM job queue. It also creates another script class_merge.sh in the Output/ directory to combine all the output files in the correct order once all the simulations (jobs) have been completed successfully. A flowchart of the script workflow and auxiliary files needed is depicted in the Figure below. File names in bold refer to existing files in the directory needed for processing
Example
To perform the HPC GSA of the "Gluttons & Muttons" java simulation program used in the example follow these steps:
1. Change directory to base/ in your class directory.
2. Test the java code from the command line to make sure it runs OK. It should finish in about 3-4 min and outputs should written to output/. Inspect input and output files (see README_PredPrey.txt for description). The first three lines below are the same as given in I. Preliminaries above and are only needed if you are starting from a new session.
> module load ufrc
> srundev -t 120
> module load java
> java -jar PredPrey1.jar -gsua input.dat output
3. If the above works (check outputs with a previous run to see if they match), test the unix script (sens_class). As explained above in section III, the script will read new inputs from the command line, substitute them into the model input file, run the java program and save the outputs to output.all. Check the outputs again to see if it worked as expected.
> sens_class 3.1 1.0 0.2 1.0 1.0 0.1 0.1 6.0
4. Now let's create the HPC jobs to run a typical GSA problem. Change directory down to your class directory.
> cd ..
5. We will start with the Morris input sample matrix (morris_eSU_matrix.txt), sampled with the eSU GSA screening matlab tool for this problem (see morris_eSU_matrix.txt and the originating PreyPred.fac input factor file in the class ZIP package distributed at the begining of this web page). In this case we have 144 simulations (rows in that file) to run. Let's copy that matrix to another file matrix.txt used in the following steps.
> cp morris_eSU_matrix.txt matrix.txt
6. Edit the class_GSA.job.top by:
a) change where it says
“/home/carpena” to “/home/YOURGATORLINK”
"carpena@ufl.edu" to "YOURGATORLINK@ufl.edu"
"/ufrc/carpena/carpena" to "/ufrc/abe6933/YOURGATORLINK/"
Notice that /ufrc/abe6933/YOURGATORLINK/ will be the "scratch" ($TMP_DIR) directory for executing the HPC jobs.
b) check that the time you are requesting is enough to execute the number of simulations in each job (be generous!).
c) save the file!
7. In this case, Since every simulation run takes ~5 min, with 144 simulations it would take ~720 min (12 h) to run this problem. We wish to run this problem in ~30 min using the HPC. I will assign 6 simulations to each HPC job and will run 24 jobs in parallel (24*6= 144 simulations). This is an important step and Video 4 below gives additional details on how to properly select the number of simulations per job based on desired execution time and available resources for your account.
8. Let's create the 24 HPC jobs with the script make_jobs.sh
> sh make_jobs.sh 144 6
9. Submit the jobs with the submission script created by make_jobs.sh
> sh class_submit.sh
10. Now wait!! To check progress ("squeue -u YOURGATORLINK"), the jobs will show as Q or PD while waiting and R when running. You will receive emails as they complete. Results are in your /home/YOURGATORLINK/class_GSA/Outputs directory.
11. Once the jobs are completed, we need to merge the output files in the correct order, from 1 to 24 so we end up with 144 rows or simulations in the final file. Change to the Outputs directory and run the script class_merge.sh created by make_jobs.sh. This will create the final output file outputs_all.txt to post-process the Morris results.
> sh class_merge.sh
12. With the combined output file from all the HPC runs (output_all.txt) continue with the postprocessing using the eSU GSA screening matlab tool for calculating Morris indices and plots.
Instructional videos
Below are step-by-step instructional videos developed by doctoral student Victoria Morgan for the Advanced Biological Systems Modeling class. They present a typical GSUA application using the scripts.
Video 1 - How to create an input factor file (.fac) for GSUA
Video 2 - How to create GSUA sample (.sam) and matrix.txt files
Video 3 - How to organize Files on the HPC
Video 4 - How to run GSUA on the HPC
Program License
These software is distributed as Freeware/Public Domain under the terms of GNU-License. If the program is found useful the authors ask that acknowledgment is given to its use in any resulting publication and the authors notified.
- Dr. Rafael Muñoz-Carpena
Professor, Hydrology & Water Quality, Biocomplexity Engineering
Department of Agricultural & Biological Engineering
University of Florida
P.O. Box 110570
287 Frazier Rogers Hall
Gainesville, FL 32611-0570
(352) 294-6747
(352) 392-4092 (fax)
carpena@ufl.edu
© Copyright 2019 - Rafael Muñoz-Carpena
References
- Muñoz-Carpena, R. Class notes on UF eLearning system.
- Alberts, J. (2017). Sim2D. Java Tutorials. Retrieved from University of Washington - Center for Cell Dynamics.
This page was last updated on June 24, 2020.